Nucleic Acid Labeling
Nucleic acid labeling is a precise molecular technique that involves attaching detectable tags to DNA, RNA, or oligonucleotides for use in various biological assays. It plays a crucial role in tracking, identifying, and quantifying nucleic acids within complex biological systems, making it essential for applications like gene expression analysis, diagnostics, and drug discovery. As an expert biotech service provider, we offer nucleic acid labeling solutions with the highest level of accuracy and customization, ensuring reliable results for advanced research and clinical applications.
What is Nucleic Acid Labeling?
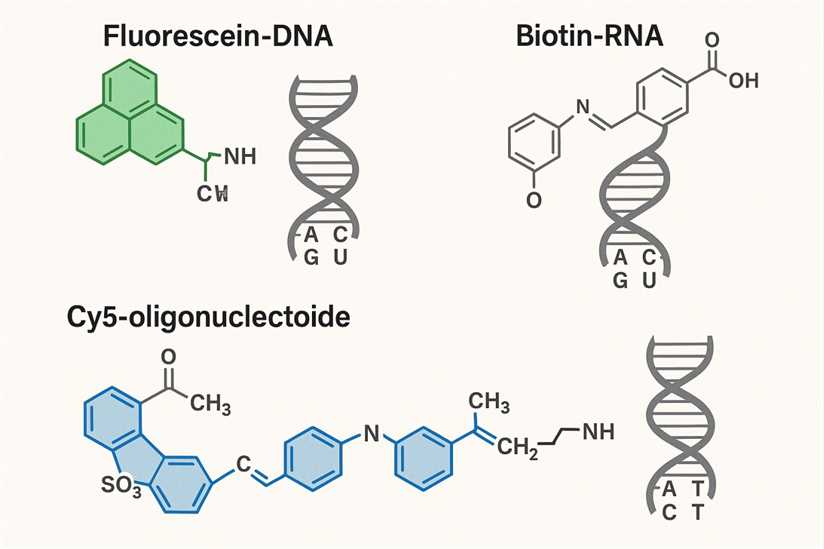
Nucleic acid labeling refers to the process of labeling or labeling DNA or RNA in molecular biology and biological research in order to track and detect these molecules during experiments. The purpose of labeling is not to affect the function of nucleic acids and research results. Here's a detailed explanation of nucleic acid labeling:
Nucleic Acid Labeling Methods: Nucleic acid labeling is mainly divided into two categories: chemical labeling and enzyme reaction-based labeling. One is chemical labeling, which uses specific chemical reagents to covalently bind labeled molecules (such as fluorescent molecules, biotin, digoxin, etc.) to nucleic acids. This method does not require the involvement of enzymes, and the reaction process is simple and efficient, and has little impact on the structure and function of nucleic acids. The second labeling method is based on the labeling method of enzymatic reaction, which is an enzymatic reaction in which label-modified nucleic acids are added to the reaction system. This method is often used in specific molecular biology experiments, such as gene editing and RNA interference experiments.
Detection Methods: Labeled nucleic acids can be detected in two ways: one is direct detection, which can be detected directly because the labeled nucleic acid may contain a reporter molecule that is used for optical, luminescent, or fluorescent signaling. Another detection modality is indirect detection, where labeled nucleic acids carry specific labeled molecules, such as biotin or digoxin, that can bind to specific reporter-coupled molecules, such as streptavidin or specific antibodies, to amplify and detect the signal.
Our Nucleic Acid Labeling Services
We offer tailored DNA labeling solutions to meet diverse research and development needs. Our services include various labeling methods, such as fluorescent, radioactive, and biotin labels, to enhance detection sensitivity and support applications in fields like genomics, diagnostics, and drug discovery.
Each labeled nucleic acid product undergoes rigorous quality control to ensure high purity, specificity, and reproducibility. Our QC process includes mass spectrometry, HPLC, and functional assays, delivering reliable and accurate results tailored to research and clinical requirements.
Our customized RNA labeling services are designed to accommodate specific project requirements, with options for various labeling techniques that facilitate high-quality results in RNA sequencing, gene expression analysis, and molecular diagnostics. We ensure optimized labeling strategies to maximize assay performance and sensitivity.
For oligonucleotide research, we provide comprehensive labeling solutions to support applications in gene editing, hybridization studies, and diagnostic assay development. Our services cover a wide range of oligonucleotide types, including siRNA, aptamers, antisense oligonucleotides (ASO), and microRNA, offering flexible options for modifying the oligo backbone, terminal ends, and internal bases. This customization enhances detection capabilities and supports specialized research needs in functional genomics, therapeutic development, and molecular diagnostics.
Our services cover a wide range of oligonucleotide types.
RNA molecules are covalently or non-covalently attached to gold nanoparticles for enhanced delivery, stability, or detection applications.
siRNA is chemically conjugated to antibodies to enable targeted gene silencing via receptor-mediated delivery.
Functional peptides are linked to RNA strands to improve cellular uptake, targeting, or intracellular trafficking.
Cholesterol is attached to siRNA to enhance membrane permeability and improve in vivo delivery efficiency.
N-acetylgalactosamine (GalNAc) is conjugated to siRNA to enable targeted delivery to hepatocytes via ASGPR-mediated uptake.
siRNA is labeled with dyes, biotin, or other tags for tracking, quantification, or affinity-based applications.
DNA strands are functionalized on gold nanoparticles for biosensing, diagnostics, or nanotechnology-based gene delivery.
DNA oligos are immobilized on Luminex beads for high-throughput multiplex detection and quantification.
DNA sequences are covalently linked to proteins for applications in biosensors, nanostructure assembly, or hybrid biomaterials.
DNA is chemically attached to antibodies for use in immuno-PCR, DNA barcoding, or antibody-guided targeting systems.
Nucleic acid binding proteins regulate DNA and RNA processes, playing essential roles in gene expression, replication, repair, and biotechnology-driven molecular diagnostics.
From Inquiry to Completion: Our Service Flow
Advantages of Working with Us
We utilize cutting-edge nucleic acid labeling technologies, including fluorescence, biotin, radioactive, and isotope labeling, ensuring high precision, sensitivity, and versatility across a wide range of applications.
We offer fully customizable labeling services to accommodate diverse research and clinical needs. Whether it's choosing the optimal label, adjusting labeling conditions, or targeting specific nucleic acid sequences, we provide flexible solutions.
We maintain stringent quality control measures throughout the labeling process, ensuring high-purity, accurately labeled nucleic acids with consistent and reproducible results.
We offer detailed reports with every project, including complete characterization of labeled nucleic acids, experimental conditions, and expected performance in downstream applications, ensuring you have all the information needed for successful integration.

Whether you require small-scale labeling for preliminary studies or large-scale labeling for clinical trials, we offer scalable solutions to meet the demands of your research or production pipeline.
Where Our Nucleic Acid Labeling Are Used?
The applications of nucleic acid labeling mainly include the following aspects:
In vitro experiments, labeled DNA or RNA can be transfected into mammalian cells. In this way, subcellular localization and target gene function can be monitored in the same experiment. This is very helpful for studying the precise location and functional status of genes.
In animals, labeled nucleic acids can be fed into mammalian cells, allowing monitoring of subcellular localization and reporter gene expression. This is very important for studying the specific role and regulatory mechanisms of genes in living organisms.
Labeled DNA or RNA samples are optimally sensitive and stable when performing hybridization reactions. The labeled DNA or RNA can be thermally denatured as needed for the hybridization application, which is critical for subsequent molecular biology experiments.
For labeled RNA samples, the RNA can be denatured by heating at 55-65 ℃ for 10 min, and then the labeled RNA probe can be treated with denaturing reagent to maintain its optimal performance in the hybridization reaction.
Case Study
Case Study 1
 Fig.1 Application of mgSrtA-mediated cell labeling in multiplexed scRNA-seq. (Liu, Yingzheng, et al., 2024)
Fig.1 Application of mgSrtA-mediated cell labeling in multiplexed scRNA-seq. (Liu, Yingzheng, et al., 2024)
Transpeptidase A (SaSrtA) in Staphylococcus aureus is able to localize a variety of bacterial proteins to the surface of bacteria through a process called transpeptidization reaction, mediating bacterial invasion of host cells and bacterial biofilm formation. In addition to important biological functions, the SaSrtA-catalyzed peptide transfection reaction is a widely used and engineered protein engineering tool that can link a protein containing a pentapeptide sequence (LPXTG) at the C-terminus to another protein with an oligoglycine chain at the N-terminus. The research team first engineered SaSrtA to obtain mgSrtA, which was found to have the ability to label nucleic acid molecules onto the surface of mammalian cells. Experiments were performed using DNA oligonucleotide strands containing G bases, and mgSrtA was verified to label different cells, including immortalized cells and primary cells, under mild conditions. Through a series of biochemical experiments and point mutagenesis analysis, the results showed that the binding of mgSrtA to nucleic acids did not depend on the traditional transpeptidase activity, and Cu²⁺ could significantly enhance its binding ability. CRISPR genome-wide screens further revealed that glycosaminoglycans (GAGs) on the cell surface play an important role in the labeling process. Flow cytometry and imaging experiments showed that mgSrtA and nucleic acid molecules colocalized on the cell surface after labeling, confirming that mgSrtA played a bridging role during labeling. Studies have also shown that mgSrtA-tagged nucleic acid molecules are able to enter the interior of cells and express proteins, providing evidence of their potential as nucleic acid delivery tools. This discovery opens up a new avenue for sample labeling in single-cell sequencing as well as the development of nucleic acid drug delivery tools.
Case Study 2
In genetics and oncology, fluorescence in situ hybridization (FISH) is widely used to detect structural variations and abnormalities in chromosomes. Using fluorescently labeled DNA probes, researchers target specific regions of the chromosome to visualize copy number changes, translocations, deletions, or duplications in these regions. In a study used in the diagnosis of breast cancer, FISH was used to detect the amplification of the HER2 gene, an important gene associated with breast cancer prognosis. When the HER2 gene is amplified, patients are often susceptible to drugs that target HER2, such as trastuzumab. The labeled DNA probe targets the HER2 gene sequence, and the fluorescent signal of the gene amplification is observed by fluorescence microscopy. With FISH technology, researchers were able to definitively detect the presence or absence of HER2 gene amplification. This method provides a reliable molecular diagnostic basis for the personalized treatment of breast cancer patients, guides the use of targeted drugs, and improves the treatment effect.
Frequently Asked Questions (FAQ)
Nucleic acid labeling is the process of attaching detectable markers, such as fluorescent dyes, biotin, or radioactive isotopes, to DNA or RNA. This allows researchers to track, visualize, or quantify specific nucleic acid sequences in various biological and medical studies, including gene expression analysis and molecular diagnostics.
Nucleotides can be labeled enzymatically or chemically by incorporating labeled molecules, such as fluorescent dyes or biotin, during DNA or RNA synthesis. Enzymes like polymerases or terminal deoxynucleotidyl transferase (TdT) are commonly used to introduce labeled nucleotides into DNA or RNA sequences for detection.
DNA labeling methods include fluorescent labeling for visualization, radioactive labeling for sensitive detection, biotin labeling for affinity binding, and enzymatic methods using polymerases to incorporate labeled nucleotides during synthesis. Chemical modifications can also be employed for post-synthesis attachment of detectable tags.
A nucleic acid-based identification method uses labeled DNA or RNA probes to detect complementary sequences in a sample. Techniques like PCR, FISH, or microarrays utilize these probes to identify specific genetic sequences, enabling the detection of pathogens, mutations, or gene expression levels in research and diagnostics.
Nucleic acid labeling is used in gene expression analysis, molecular diagnostics, in situ hybridization, and protein-nucleic acid interaction studies. It enables visualization and quantification of DNA or RNA sequences in cells, tissues, or sequencing experiments, playing a key role in research and diagnostic applications.
Nucleic acid labeling is essential in sequencing, as it allows for the detection of DNA or RNA fragments. In Sanger sequencing, labeled nucleotides help visualize DNA synthesis, while in next-generation sequencing (NGS), labeled primers or adapters facilitate large-scale detection of sequences for genomic or transcriptomic analysis.
Learn More About Nucleic Acid Labeling
References
- Liu, Yingzheng, et al., The Transpeptidase Sortase A Binds Nucleic Acids and Mediates Mammalian Cell Labeling. Advanced Science (2024): 2305605.
- Bartley, Angela N., et al., HER2 testing and clinical decision making in gastroesophageal adenocarcinoma: guideline from the College of American Pathologists, American Society for Clinical Pathology, and American Society of Clinical Oncology. American journal of clinical pathology 146.6 (2016): 647-669.